Exploration of RNA-Binding Proteomes
Using RNA as a handle UV-crosslinked proteins can be co-purified and detected by MS. Previously, this was only possible for polyadenylated RNA, which was extracted and purified from total lysates with poly(dT) beads - a procedure termed interactome capture [Castello et al., Cell, 2012; Baltz et al., Mol Cell, 2012]. XRNAX extracts can be used as a a starting point for the purification of protein-crosslinked RNA using conventional silica columns, thereby giving insights into the complete RNA-binding proteome independent of polyadenylation.
Learn here about the protocol for the discovery of RNA-binding proteins starting from XRNAX extracts (refer to The Protocol section for initial XRNAX extraction).
XRNAX
For the controlled discovery of RNA-binding proteins use 1000 µg of XRNAX extract (2 confluent 245 x 245 mm2 dishes MCF7 cells yield approximately 1000 µg XRNAX extract). Expand cells with a heavy and a light SILAC label until confluent, UV-crosslink and harvest cells with one SILAC label as described in the ‘The Protocol’ section. Harvest cells with the other SILAC label without UV-crosslinking. Combine crosslinked and non-crosslinked cells during the initial TRIZOL lysis step of XRNAX. Proceed with the XRNAX extraction without further changes.
Downstream Protocol for the Exploration of RNA-Binding Proteomes
Required Material
Tris-Cl 1 M (pH=7.5)
DTT 1 M
SDS 20 %
Trypsin/LysC (Promega, V5073)
Chloroacetamide (CAA) 1 M
GlycoBlue (Ambion, AM9515)
NaCl 5 M
Quiagen RNeasy Midi Kit
Ethanol 100 %
TEAB 20 mM (pH=8.0)
RNase A (Thermo, EN0531)
RNase I (Ambion, AM2295)
RNase T1 (Thermo, EN0541)
High pH buffer 10 x (ammonium formate 200 mM, pH=10)
Formic acid 1 %
Partial Tryptic Digestion
To 950 µg of XRNAX extract add 50 µl tris-Cl 1 M, 5 µl SDS 20 %, 10 µl DTT 1 M and mix. Reduce cysteins at 60 °C for 30 minutes, 700 rpm shaking. Subsequently, allow samples to reach room temperature. Add 20 µl CAA 1M, mix and allow alkylation to occur for another 30 minutes at room temperature. Add 100 ng trypsin/LysC and allow partial digestion to occur for 15-30 minutes. Stop the partial digestion by combining the sample with 3.5 ml buffer RLT in a 15 ml falcon tube (refer to Quiagen RNeasy Midi Kit manual for buffer descriptors) and mix by inversion.
Silica Purification
For the purification of protein-crosslinked RNA from the partially digested XRNAX extracts use the Quiagen RNeasy Midi Kit with modified protocol. Heat the partially digested XRNAX extract in buffer RLT to 60 °C for 15 min. Let the sample reach room temperature and add 2.5 ml of ethanol 100 %. In the following the same sample will be purified over the same RNeasy Midi spin column four times. This way all protein-crosslinked RNA is extracted, while only one column is required instead of four. Mix by inversion and apply two times 3.5 ml sample to a spin column by centrifugation with 3000 g for 3 minutes. Do not discard the flow-through but transfer it to a 15 ml tube for storage. Wash the column twice with 2.5 ml buffer RPE by centrifugation with 3000 g for 1 minute. Do not use buffer RW1. Elute the protein-crosslinked RNA with 250 µl nuclease-free water by centrifugation with 3000 g for 3 minutes into a 15 ml tube. Use this tube to collect all eluates from the 4 identical rounds of purification. All eluates should combine to approx. 900 µl, which are transferred to a fresh 2 ml tube. Add 60 µl NaCl 5 M along with 1 µl glycoblue and 1 ml isopropanol. Mix by inversion and incubate for 1 hour at -20 °C. Spin down with 18000 g (or full speed) at -10 °C (or coldest setting) for 60 minutes. Discard the supernatant and wash the pellet with 2 ml of 70 % ethanol. Remove all residual ethanol and take up the pellet in 45 µl TEAB 20 mM.
RNase Digestion
Heat the sample to 85°C for 5 minutes and cool on ice before adding 1.5 µl of RNase A, RNase I and RNase T1. Allow RNA digestion to occur for 12 hours at 37 °C, 700 rpm shaking.
Complete Tryptic Digestion
Add 500 ng trypsin/LysC in 5 µl resuspension buffer (Promega) and digest for 16 hours at 37 °C, 700 rpm shaking.
High pH Fractionation
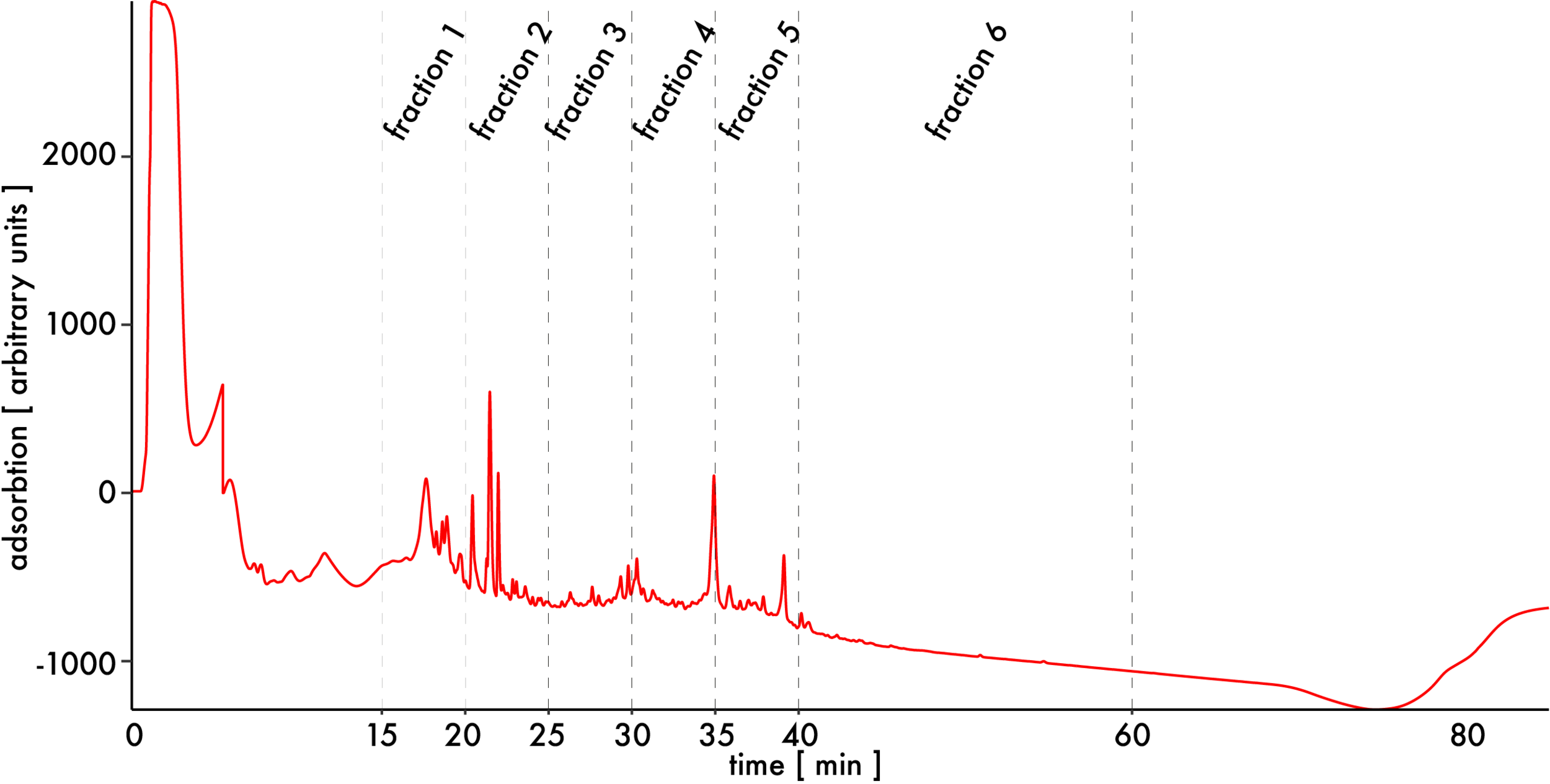
We fractionate samples using a 20 mM ammonium formate buffer system at pH=10. Therefore add 20 µl of 10 x high pH buffer and spin down the sample for 5 minutes at 20000 g. Transfer the supernatant to an injection vial and fractionate. Alternatively, preclean the sample using stage-tip or Oasis peptide cleanup. This might help with sample impurities, which usually arise from incomplete RNase digestion or detergents in the RNase storage buffers. Note that with the amounts of RNase and the high pH chromatography system used in this protocol we did not encounter difficulties if RNase digestion occurred for a long enough time.
The sample shown here was fractionated with the gradient 0-2 minutes 0 % B, 2-60 minutes linear gradient to 65 % B, 61-62 minutes linear gradient to 85 % B, 62-67 minutes 85 % B, 67-85 minutes 0 % B on an Agilent Infinity 1260 LC system (Agilent) using a Phenomenex Gemini 3 µM C18, 100 x 1 mm column (Phenomenex):
Early fractions before 15 minutes contain few peptides but a lot of RNA fragments. Exclude those fractions and combine the rest into 6 consecutive ones. Completely dry samples by speedVac, take them up in 15 µl formic acid 1 % and analyze them by HPLC-MS.